Adipocyte Cell Imaging Challenge
During November 2020 AI Sweden and AstraZeneca challenged the AI community to solve the problem of labeling cell images without requiring toxic preprocessing of cell cultures. The task was to utilize machine learning to predict the content of the fluorescence images from the corresponding bright field images.

Background
Historically, most drugs have been small molecules binding to target proteins in the body. The problem is that one drug might bind to several similar proteins, creating unwanted side effects. A more specific way is to target the RNA sequence of the proteins where nanomedicines often are used to transport RNA cargo through the body. These nanomedicines are small engineered machines made from biomolecules that interact with cell membranes and machinery to deliver RNA into cells where it can affect their function.
To be able to inject the nanomedicines into the skin will make treatments with nanomedicines easier for patients. One of the most important cellular targets is the fat cells (adipocytes) that we all have in our skin. To avoid using human test subjects, adipocyte cell cultures can be created from stem cells. One of the most important methods to investigate the uptake of nanomedicines in cells is cell imaging, which is the topic of this challenge.
The goal of cell imaging is to extract relevant information about the cell structures that can guide pharmaceutical development. To permit imaging of different cell structures, fluorescence microscopy is used to label specific parts of the cell.
AstraZeneca has shared a data set consisting of pictures of stem-cell derived human adipocyte cell cultures The cells have been imaged, using a robotic confocal microscope, at three different magnifications, using both brightfield and fluorescence imaging.
The task
The task was to utilize machine learning to combine the advantages of bright field and fluorescence imaging and at the same time avoid the toxic effects of cell labeling by predicting the content of the fluorescence images from the corresponding bright field images.
Download the complete problem formulation
Data
There are three sets of images corresponding to three different magnification settings (20x, 40x, and 60x) of the microscope. For each field of view, there will be seven bright field images for different values of the focal plane, and three different fluorescence images corresponding to the labeling of nuclei, lipocytes, and cell-matrix respectively. There will be on the order of 50-100 images for each magnification setting. Each image is approximately 2156 by 2556 pixels in size, using 16 bits to represent each pixel value.
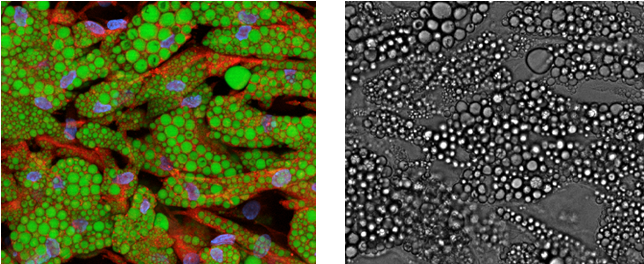
To the left is a superimposed fluorescence image of cell nuclei (blue), lipid droplets (green), and cytoplasm (red). Each color is a single-channel image. To the right is the corresponding bright field image.
Teams
BIOMAG
A team with researchers from Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, and Biological Research Centre of the Hungarian Academy of Sciences
BioMedAI-Lund
An interdisciplinary team with extensive expertise in deep learning and biomedical image
analysis, including the analysis of large-scale phenotypic high-content imaging screens. All team members are senior researchers at Lund University, collaborating as part of the AI Lund network.
HASTE team
The HASTE Team consists of five PhD students from Uppsala University, Sweden. The team have a range of experience in image cytometry using both traditional and deep learning approaches
NordAxon Code Monkeys
The NordAxon Code Monkeys is a team of three employees from NordAxon, a regional Machine Learning consultancy business located in Malmö.
rähmä.ai
A team of two computer vision experts from Silo AI with a background related to healthcare and medical fields.
Soft Matter Lab @ GU
The Soft Matter Lab is a research group at the Department of Physics of the University of Gothenburg. The group focuses on research in biomedical optics, biomimetic active systems, neurosciences, and machine intelligence.
The Bug Hunters
A team of Medical Engineering graduates from KTH Royal Institute of Technology. The team has a background in medical imaging and machine learning, we are highly interested in data science for healthcare.
Yenomze
A multidisciplinary team consisting of three Deep Learning graduates from Linköping University and one 2nd year master’s student in Molecular Biology and Genetics at Izmir Institute of Technology.
Evaluation
The teams were evaluated in two steps:
1. Quality metrics
A set of pre-defined quality metrics was used to measure the objective quality of the generated images.
Evaluation metrics code can be found here
2. Jury presentation & report
A jury with members from AstraZeneca, AI Sweden, and Vinnova assessed the quality of the different contributions based on the presentation given by each group. The report from each group was evaluated by AstraZeneca.
Results
Read more about the outcome of the challenge here.
Final ranking (including quality metrics, report, and jury presentation)
1. HASTE team
2. SoftMatter Lab @ GU
3. BIOMAG
4. rähmä.ai
5. NordAxon Code Monkeys
6. The Bug Hunters
7. Yenomze
8. BioMedAI-Lund
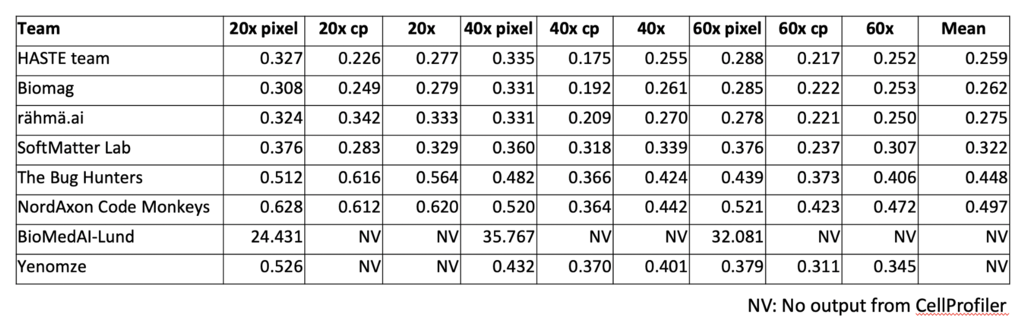
Model performance
Facts
- The competition was held online between November 2 and November 23 2020
- The prize sum, sponsored by AstraZeneca is $5000
- 8 teams participated
- The teams had access to AI Sweden's computational resources during the competition
About the Dataset